It is surprising, then, to find that many DNA-binding proteins can bind more tightly to sequences that have been engineered to contain a type of single-nucleotide change called a mismatch.
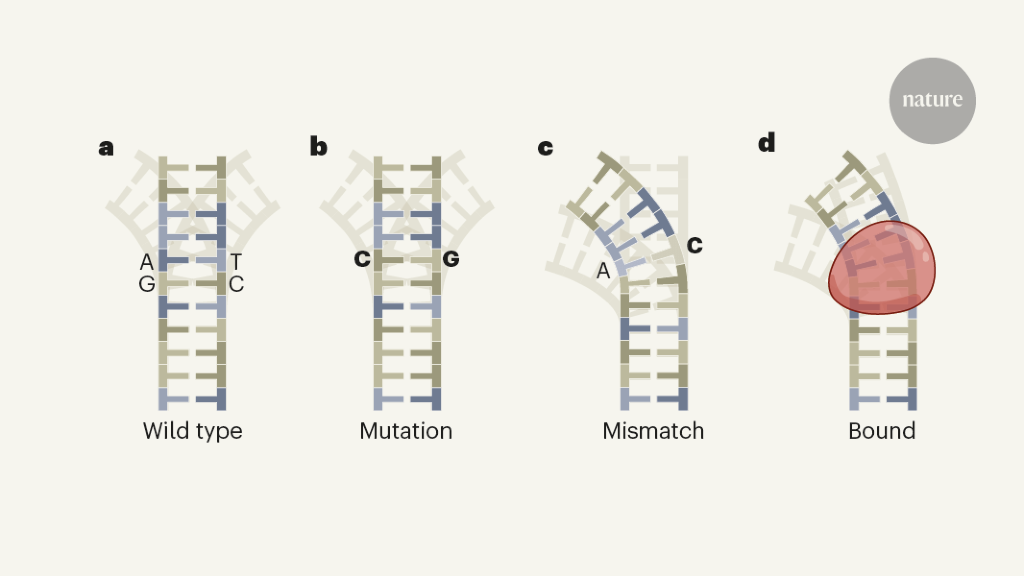
In normal base-pairing, adenine (A) bases on one strand of the DNA duplex pair with thymine (T) on the complementary strand, and guanine (G) bases pair with cytosine (C) — so a change from an A–T pair to a C–G pair is a mutation, whereas a change to A–C is a mismatch.
The DNA double helix involves pairs of DNA bases: adenine (A) bases on one strand pair with thymine (T) on the other, and guanine (G) bases pair with cytosine (C).
b, A mutation changes a pair of bases into another pair (such as A–T to C–G).
d, Binding by proteins (red) can also alter DNA conformations.
In simple terms, shape readout is the ability of proteins to indirectly recognize specific DNA sequences by their characteristic 3D shapes2,3.
This is in contrast to their ability to directly recognize specific sequences by the characteristic chemical groups present in each base pair, a mechanism known as base readout.
Taking advantage of this, a protein that needs to bind to a specific sequence can try to bend any sequence it encounters in a way that would be most compatible with its intended target.
realized that mismatches, being small changes to a sequence that cause large changes in shape, offer a unique way to study this phenomenon.
The authors therefore developed what they call a saturation mismatch-binding assay (SaMBA), which quantifies the binding of a protein to every possible single-nucleotide mismatch in a particular DNA sequence.
Briefly, they manufactured a microchip arrayed with single strands of DNA encoding every possible single-nucleotide variant of a consensus sequence.
The complementary DNA hybridized with each of the strands printed on the chip, creating duplex DNA with every possible mismatch.
SaMBA revealed not only that it is possible for mismatches to improve DNA–protein binding, but also that it is relatively common for them to do so.
For some proteins, the most effective mismatch occurred in the natural target sequence, making the protein bind to that sequence even more tightly.
For others, the most effective mismatch occurred in a non-target sequence, and made the protein bind to that sequence at levels comparable to those of the natural target.
In both cases, the same mechanism is predominant: the mismatch pays the energetic cost of distorting the DNA so that the protein doesn’t have to.
Note that to actually improve binding, the mismatch must distort the DNA in the same way as the protein would through the shape-readout mechanism.
The mismatch also should not interfere with any chemical contacts between the protein and the DNA — although the authors did find that mismatches can sometimes introduce favourable contacts.
In future, perhaps nucleotides that do not occur in nature4 could be used in SaMBA to thoroughly probe the array of conformations that DNA can adopt, similarly to the way in which unnatural amino acids have been used to investigate subtle changes in protein biophysics5.
More broadly, the finding that mismatches often improve binding might have implications for diseases such as cancer.
But the clear propensity for mismatches to improve binding makes such a mechanism worth contemplating.